I’ve started having even more fun with Kotlin. One way you can learn Kotlin is by practicing and solving real exercises. That’s how I discovered Exercism.org, a free way to learn dozens of languages by jumping head first into problems.
Love how they probably combined the words “exercise” and “exorcism” to come up with this branding and it helped, the name sticks with you. Here’s what I like about Exercism:
- you have exercises for the most well-known programming languages (including Kotlin).
- it’s free to use
- you submit your solutions and then receive feedback from other users thus you are helped to learn by other peers
Anyway, maybe in a future blog post I’ll give more details on how Exercism helps with learning a new programming language. In the meantime, I had a lot of fun solving one of their problems called “Hamming”
The Hamming Distance problem in Kotlin
Here’s an overview of what the problem was about:
You know how every living matter contains cells and those cells have DNA strings in them. Each cell in living organisms divides and gives birth to other cells inheriting DNA. And that happens a lot, especially for the human body. In average, during a lifetime, a human body will have over 10 quadrillion cell divisions.
With numbers so large, it’s obvious that things will also go wrong. When a cell divides into other sub-cells, the parent DNA will be replicated onto the child cells. But with high numbers like this, mistakes happen and some DNA strings get encoded with incorrect information. Usually when a cell divides, the child should have the same exact DNA string. However, if you compare two strands of DNA and count the differences, you’re actually counting how many mistakes occurred. The number of mistakes (i.e. differences between DNA strings) is also known as the “Hamming Distance”.
When we read a DNA string we use the letters C,A,G and T (thymine (T), adenine (A), cytosine (C), and guanine (G)). So two strands of DNA might look like this:
AGCTGACCTAACGGGAT
GATCGGTCAGACGGCCT
^^^^-^^-^^^---^^-The two strings above have 11 differences, and thus the Hamming Distance is 11. Not just biology uses Hamming Distance as way to express differences, other fields in science use it so it’s good to know how to calculate it.
So the problem is this, try to write a function in Kotlin that calculates the Hamming Distance between 2 DNA strings given as arguments. The Hamming Distance can be calculated only by comparing 2 sequences of equal length, if the strings aren’t equal an exception should be raised.
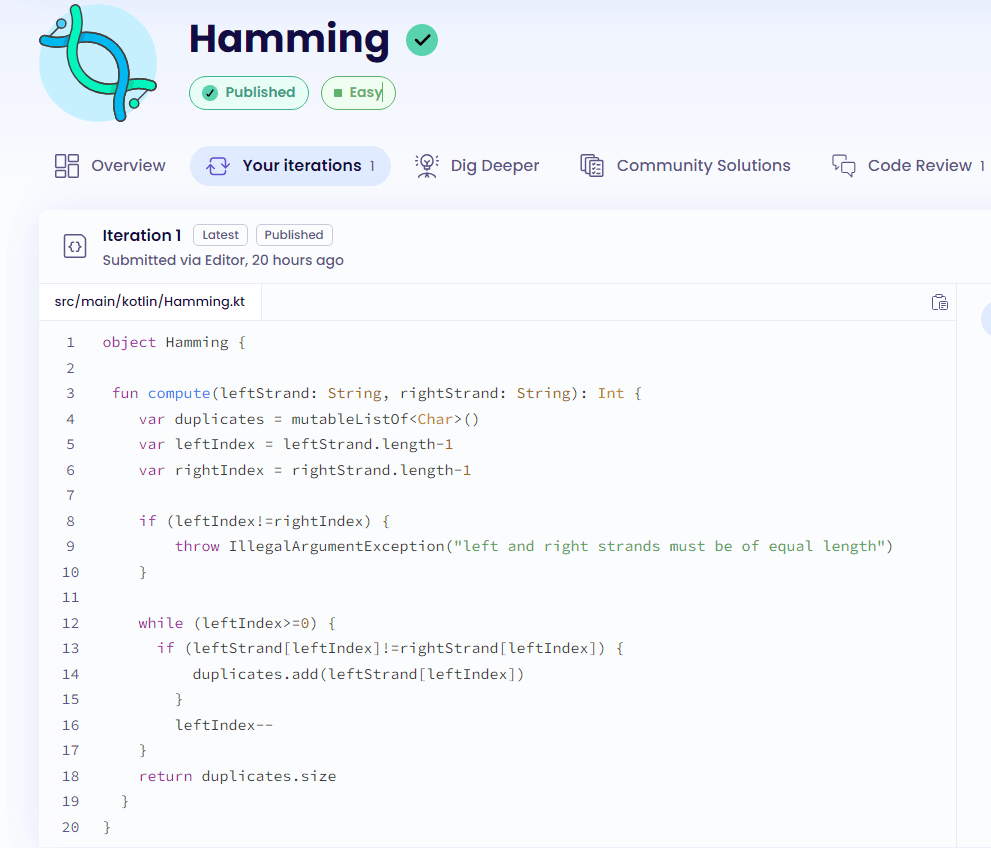
My solution to this problem is a bit rudimentary (too rudimentary), because I didn’t get (yet) too far on the learning Kotlin path. But it’s still a valid solution, and below you can see how to calculate the Hamming Distance using Kotlin:
object Hamming {
fun compute(leftStrand: String, rightStrand: String): Int {
var nonDuplicates = mutableListOf<Char>() //i'm creating a list where I'll save the characters that are different
var leftIndex = leftStrand.length-1 //counting the lenght of the strings to use for iterations and checks
var rightIndex = rightStrand.length-1
//checking if the strings are differnt and throwing an error if they don't match
if (leftIndex!=rightIndex) {
throw IllegalArgumentException("left and right strands must be of equal length")
}
/*this is where fun happens, i'm looping through the whole string and comparing characters. If they don't match, i save those in my nonDuplicates list*/
while (leftIndex>=0) {
if (leftStrand[leftIndex]!=rightStrand[leftIndex]) {
nonDuplicates.add(leftStrand[leftIndex])
}
leftIndex--
}
return nonDuplicates.size //in the end I just return the size of the list I used to save differences in
}
}
Obviously the approach could’ve been prettier, but here was my thought:
- instead of counting differences with a counter, I decided to save each character that is different in a list, because I wanted to see what it saves to be sure I did it right
- each string in Kotlin (and many other languages) is basically a list of Char elements, thus you can check each character in it through iterations
- in the end, after my list of nonDuplicates is all filled up, I simply count how many elements it has and that’s the humming distance which I return for the compute method.
Anyway, as mentioned above is a quite rudimentary solution to the problem. One of the most elegant and short solutions in Kotlin for the Humming Distance is this one:
object Hamming {
fun compute(s1: String, s2: String): Int {
if (s1.length != s2.length) throw IllegalArgumentException("left and right strands must be of equal length.")
return s1.zip(s2).count { it.first != it.second }
}
}
As you can see, it only has a few lines of code thus much more efficient than my solution. Here’s what it does different:
- it’s associating the strings sent as arguments for the compute method to s1 and s2.
- it checks to see if the lenghs of the 2 strings match, throwing an IllegalArgumentException if they don’t.
- lastly, it uses the zip function combined with count to see if the characters in one string, are different from the ones in the other string.
The zip function is used to create sequences of Pair values of the characters at the same index in both input Strings.
Since zip is marked as an infix function, it can be called between the two strands (i.e. between the left receiver and the right parameter), as well as using the second one as parameters.
Both these declarations are valid:
leftStrand.zip(rightStrand)
leftStrand zip rightStrandLet’s consider two short strings as an example:
AGCA
GATA
If we were to println the expression s1.zip(s2) this is what we’d get, a list of pairs:
[(A,G), (G,A), (C,T), (A,A)]Now that we have the pairs of characters, it’s time to count the different ones. The count function iterates the Pair values and uses the it keyword to count the Pair values that meet the Boolean condition of the first value in the current Pair not being equal to the second value in the Pair. Don’t be scared of the “it” keyword, basically it’s a temporary value used to iterate through the list.
At the end, the result of count (which is an integer) will be returned.
Again, both solutions passed the given tests, though the second one is much more elegant. I prefer mine because I was able to troubleshoot it better by displaying saved characters that were different. Anyway, have fun with Kotlin!